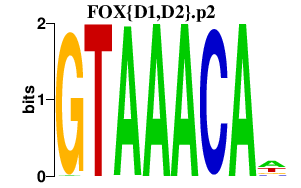
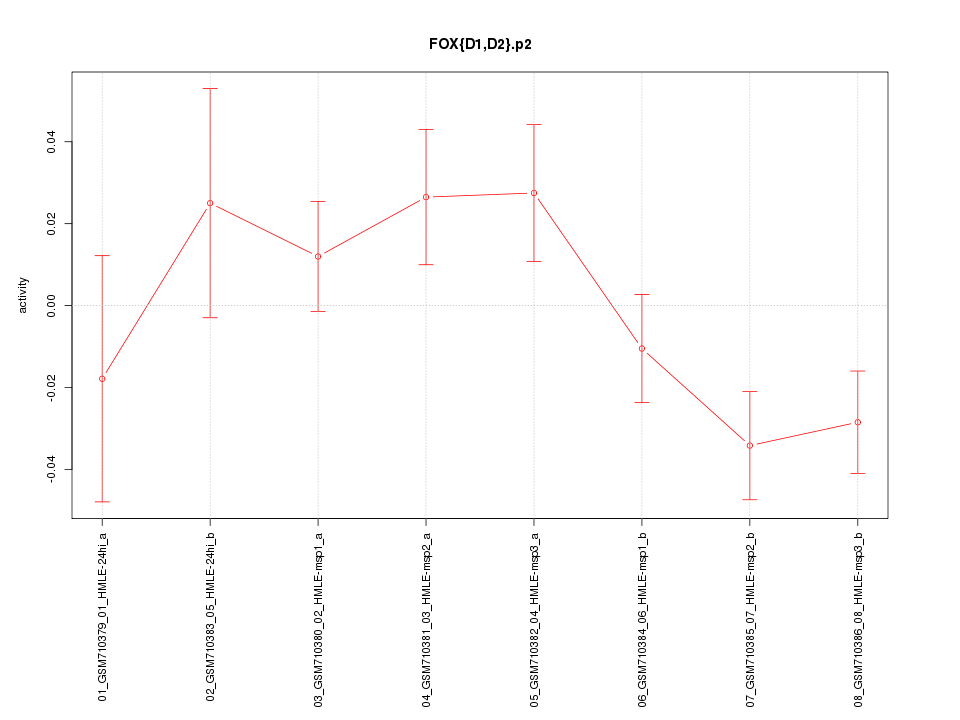
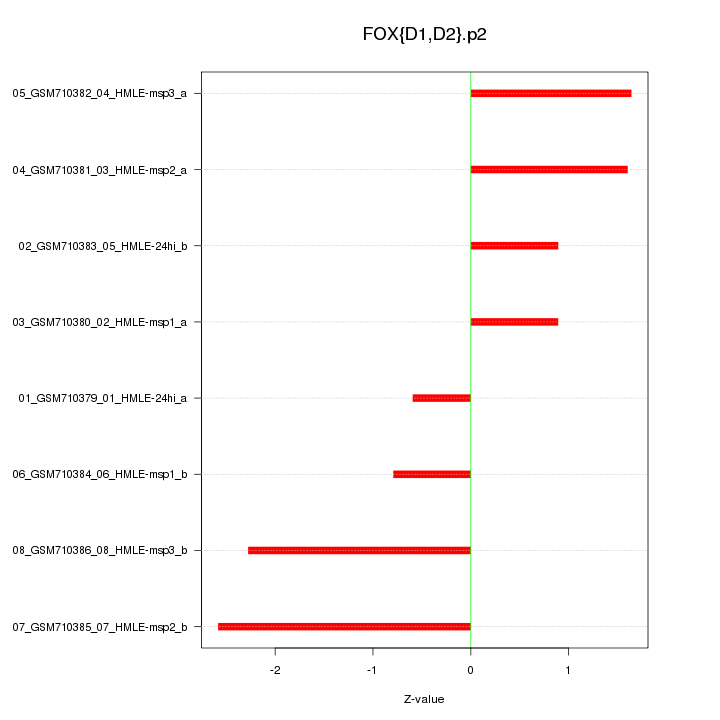
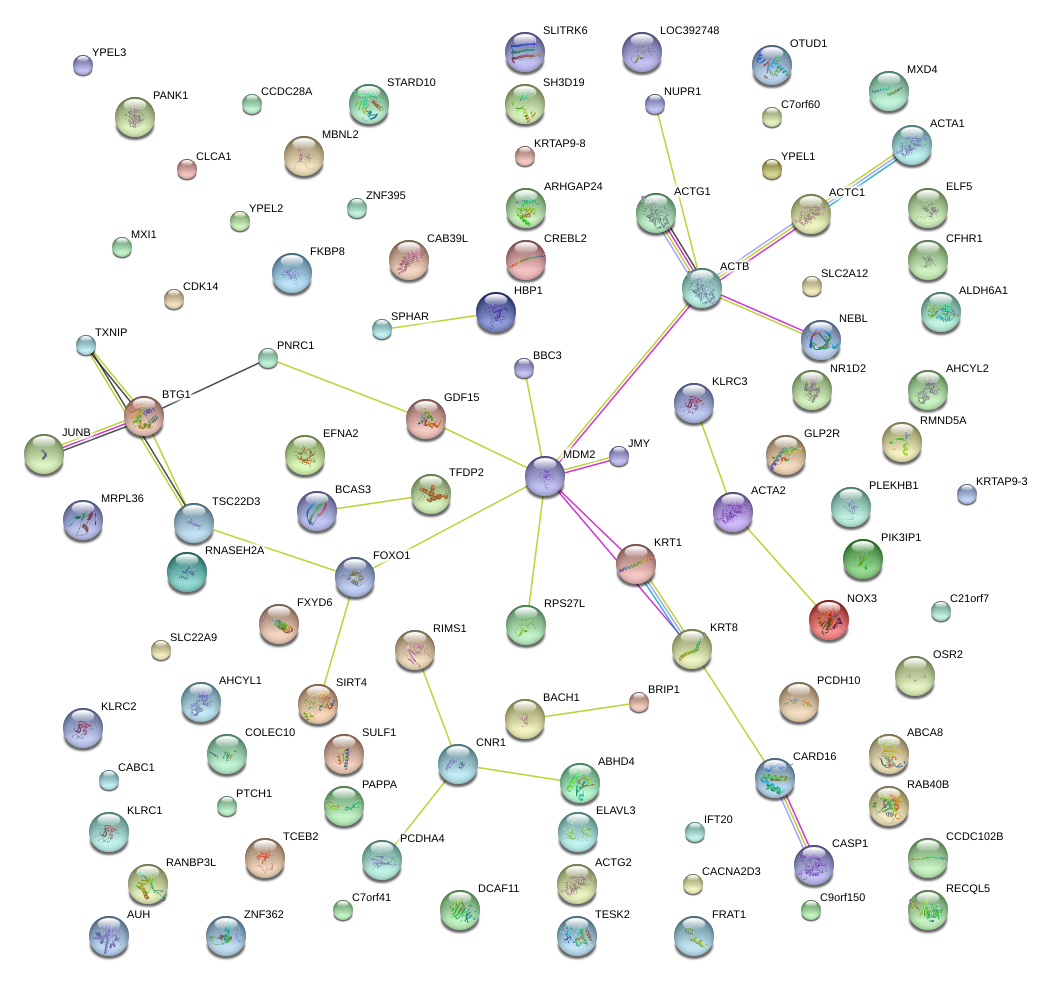
|
chr10_+_111985628
|
1.322
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chr10_+_111969988
|
1.315
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr17_+_58755158
|
1.014
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr18_+_66465316
|
0.956
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr6_+_89791510
|
0.927
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr19_+_18496966
|
0.838
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr12_-_92539614
|
0.834
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr17_+_9745847
|
0.709
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr6_-_88875766
|
0.660
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr11_-_72504643
|
0.646
|
NM_006645
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10
|
|
chr10_+_23728197
|
0.640
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr16_-_30107492
|
0.624
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr11_-_104916041
|
0.612
|
NM_001017534
NM_052889
|
CARD16
|
caspase recruitment domain family, member 16
|
|
chr4_-_2263706
|
0.581
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr1_+_145438437
|
0.579
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr21_+_30517897
|
0.575
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr10_+_99079017
|
0.569
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr7_+_106809405
|
0.567
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr9_-_3223007
|
0.562
|
|
|
|
|
chr12_-_10573041
|
0.558
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr19_+_1207129
|
0.556
|
|
EFNA2
|
ephrin-A2
|
|
chr17_-_26662439
|
0.537
|
NM_174887
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas)
|
|
chr13_+_97874704
|
0.535
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr7_-_112579726
|
0.533
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr17_+_57408907
|
0.531
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr9_-_98269480
|
0.515
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr1_+_229440128
|
0.512
|
NM_006542
|
SPHAR
|
S-phase response (cyclin related)
|
|
chr7_+_30174343
|
0.505
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr17_-_66951381
|
0.501
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr9_+_12775713
|
0.501
|
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr13_-_41240607
|
0.500
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr14_-_74551073
|
0.473
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr9_+_118916069
|
0.466
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr4_+_134073180
|
0.462
|
|
PCDH10
|
protocadherin 10
|
|
chr8_+_70405027
|
0.459
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr10_-_21186530
|
0.444
|
NM_006393
|
NEBL
|
nebulette
|
|
chr19_-_47734215
|
0.442
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr14_-_74551159
|
0.440
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr19_+_12902295
|
0.438
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr22_-_31686707
|
0.431
|
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr17_-_80656461
|
0.419
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr12_-_10588543
|
0.415
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr3_-_141868208
|
0.409
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr17_+_39394269
|
0.394
|
NM_031963
|
KRTAP9-8
|
keratin associated protein 9-8
|
|
chr10_-_90750955
|
0.388
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr19_-_11564568
|
0.386
|
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr8_+_70405080
|
0.386
|
|
SULF1
|
sulfatase 1
|
|
chr3_+_23987514
|
0.384
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr15_-_63449479
|
0.382
|
NM_015920
|
RPS27L
|
ribosomal protein S27-like
|
|
chrX_-_106960028
|
0.382
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_-_106960268
|
0.382
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr9_-_94124194
|
0.380
|
NM_001698
|
AUH
|
AU RNA binding protein/enoyl-CoA hydratase
|
|
chr14_+_24584467
|
0.379
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_+_24584009
|
0.375
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr8_-_28243941
|
0.369
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr14_+_24583905
|
0.365
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_+_24583979
|
0.363
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_+_23067128
|
0.362
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr19_-_18654280
|
0.356
|
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr6_+_72922513
|
0.355
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_+_54156691
|
0.355
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr2_+_86947429
|
0.352
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr7_+_129008044
|
0.349
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr8_+_99956630
|
0.349
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr5_+_78531848
|
0.348
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr9_-_98269686
|
0.344
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr19_-_18653680
|
0.342
|
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr4_+_86396251
|
0.341
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_+_33722158
|
0.340
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr6_+_139094656
|
0.340
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr1_+_227127959
|
0.337
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr14_+_24584321
|
0.336
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr12_+_12764755
|
0.333
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr7_+_90339146
|
0.330
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr11_+_63137260
|
0.328
|
NM_080866
|
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9
|
|
chr12_-_53343607
|
0.326
|
|
KRT8
|
keratin 8
|
|
chr22_-_31688329
|
0.324
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr12_-_53074172
|
0.324
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr12_+_12764844
|
0.324
|
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr12_+_120740123
|
0.319
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr6_-_134373618
|
0.317
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr10_-_91405320
|
0.317
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr1_+_227127993
|
0.317
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr11_+_73359734
|
0.316
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr16_-_2827128
|
0.314
|
NM_007108
NM_207013
|
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B)
|
|
chr13_-_86373328
|
0.313
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr11_-_34535302
|
0.311
|
NM_001243080
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr2_+_86947570
|
0.310
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr16_-_28550227
|
0.309
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr13_-_49975734
|
0.307
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr4_-_152149042
|
0.304
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr12_+_69201979
|
0.304
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr1_-_45956795
|
0.302
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr5_-_36301999
|
0.302
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr12_+_69201845
|
0.299
|
NM_002392
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr6_-_155777036
|
0.299
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr1_+_196788860
|
0.298
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr3_+_54156573
|
0.292
|
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr11_-_117747331
|
0.292
|
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr1_+_86934525
|
0.290
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr1_+_227127999
|
0.289
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_+_227127937
|
0.287
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr21_+_30671736
|
0.286
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr8_+_120079423
|
0.284
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr9_-_88897372
|
0.281
|
NM_030940
|
ISCA1
|
iron-sulfur cluster assembly 1 homolog (S. cerevisiae)
|
|
chr20_+_54823787
|
0.280
|
NM_019888
|
MC3R
|
melanocortin 3 receptor
|
|
chr11_-_62314199
|
0.272
|
|
AHNAK
|
AHNAK nucleoprotein
|
|
chr4_-_141075232
|
0.272
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr3_-_57234279
|
0.270
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr5_-_169724614
|
0.264
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr3_+_23986735
|
0.263
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_-_114343052
|
0.261
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_+_132480063
|
0.261
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr17_+_57642885
|
0.260
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr16_-_4664894
|
0.259
|
NM_145253
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr2_+_86947387
|
0.257
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr20_-_1309809
|
0.256
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr19_+_18111923
|
0.255
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr17_+_38219040
|
0.254
|
NM_001190918
NM_003250
NM_199334
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr2_-_26205437
|
0.251
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr19_+_50380681
|
0.248
|
NM_001168222
NM_024682
|
TBC1D17
|
TBC1 domain family, member 17
|
|
chr19_-_3061245
|
0.247
|
|
AES
|
amino-terminal enhancer of split
|
|
chr17_-_39165343
|
0.243
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr6_+_122793061
|
0.241
|
NM_181794
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta
|
|
chr1_-_198510074
|
0.237
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chr20_-_34241830
|
0.237
|
NM_003915
|
CPNE1
|
copine I
|
|
chr10_+_48255223
|
0.236
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr1_+_150122095
|
0.235
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr14_-_73493744
|
0.235
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr5_+_40841296
|
0.234
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chrX_+_70315637
|
0.233
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr14_-_76448091
|
0.233
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr12_+_6875686
|
0.232
|
|
PTMS
|
parathymosin
|
|
chr2_+_12856813
|
0.229
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr14_-_65569056
|
0.227
|
|
MAX
|
MYC associated factor X
|
|
chrX_-_48937457
|
0.227
|
|
WDR45
|
WD repeat domain 45
|
|
chrX_-_62780748
|
0.226
|
|
LOC92249
|
uncharacterized LOC92249
|
|
chr12_-_9268440
|
0.225
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr7_-_27135524
|
0.225
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr5_-_150460599
|
0.225
|
NM_001252385
NM_001252386
NM_001252393
NM_006058
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr12_-_31479120
|
0.224
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr14_+_74551655
|
0.222
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr14_-_65569221
|
0.221
|
NM_002382
NM_145112
NM_145113
NM_145114
NM_145116
NM_197957
|
MAX
|
MYC associated factor X
|
|
chr18_+_61575203
|
0.220
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chrX_-_48937560
|
0.220
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr12_-_7245048
|
0.219
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr20_+_62887046
|
0.217
|
NM_001104925
NM_018257
|
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2
|
|
chr1_+_150122415
|
0.217
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr5_-_150460550
|
0.216
|
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr10_-_99094427
|
0.213
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr12_-_9268513
|
0.213
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr6_-_30043547
|
0.213
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr1_-_161102367
|
0.212
|
|
DEDD
|
death effector domain containing
|
|
chr14_+_105266878
|
0.212
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr1_+_3541550
|
0.209
|
NM_182752
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chr2_-_198175458
|
0.209
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr12_-_7245006
|
0.208
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr17_+_67410825
|
0.205
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr20_+_21106623
|
0.203
|
NM_001163022
NM_001163023
NM_018474
|
PLK1S1
|
polo-like kinase 1 substrate 1
|
|
chr18_+_9475528
|
0.198
|
NM_006788
|
RALBP1
|
ralA binding protein 1
|
|
chr2_-_26205327
|
0.197
|
|
KIF3C
|
kinesin family member 3C
|
|
chr19_-_18902077
|
0.197
|
NM_000095
|
COMP
|
cartilage oligomeric matrix protein
|
|
chr10_-_45474210
|
0.196
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr17_-_36831156
|
0.196
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chrX_-_135863502
|
0.196
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_-_9268522
|
0.195
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_150122246
|
0.192
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr4_-_103682126
|
0.192
|
|
MANBA
|
mannosidase, beta A, lysosomal
|
|
chr21_+_30671217
|
0.191
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr1_-_161102457
|
0.190
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr10_+_99400469
|
0.189
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr19_-_50316360
|
0.189
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr6_-_33168163
|
0.187
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr2_-_208890207
|
0.187
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr3_-_93692898
|
0.187
|
NM_000313
|
PROS1
|
protein S (alpha)
|
|
chr10_-_90712510
|
0.186
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr17_+_3379295
|
0.186
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr3_-_93692592
|
0.184
|
|
PROS1
|
protein S (alpha)
|
|
chr19_+_859658
|
0.184
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr9_-_94124149
|
0.183
|
|
AUH
|
AU RNA binding protein/enoyl-CoA hydratase
|
|
chr1_-_161102210
|
0.181
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr7_-_140340575
|
0.181
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr1_+_151170978
|
0.181
|
NM_001135636
NM_001135637
NM_001135638
NM_003557
|
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha
|
|
chr2_-_69098502
|
0.180
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr9_-_130667571
|
0.180
|
|
LOC100131355
ST6GALNAC6
|
uncharacterized LOC100131355
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr12_-_31479038
|
0.180
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr13_-_28543143
|
0.177
|
|
CDX2
|
caudal type homeobox 2
|
|
chr5_-_40756009
|
0.176
|
NM_012382
|
TTC33
|
tetratricopeptide repeat domain 33
|
|
chr17_-_45973142
|
0.176
|
|
|
|
|
chr12_-_57704245
|
0.176
|
NM_014925
|
R3HDM2
|
R3H domain containing 2
|
|
chr21_+_30671227
|
0.175
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr15_+_81589240
|
0.175
|
NM_004513
|
IL16
|
interleukin 16
|
|
chr20_-_50179338
|
0.174
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr16_-_28550324
|
0.173
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chrX_-_136113832
|
0.173
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr14_+_24867926
|
0.171
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr1_-_1356823
|
0.171
|
NM_001145210
NM_001243535
NM_001243536
|
ANKRD65
|
ankyrin repeat domain 65
|
|
chr7_+_129007942
|
0.169
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_+_33061560
|
0.168
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr3_-_187452669
|
0.167
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|